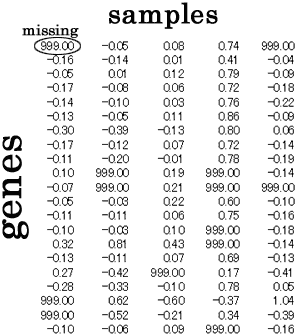
BPCAfill is a missing value estimation software suitable for gene expression profile data. (DNA microarray)
BPCAfill estimates missing values in the gene expression matrix, and fill them by the estimated values. The estimation almost always exhibits the best accuracy of all existing methods (as long as I know in 2003). Especially, when your data set has large number of genes and/or large number of samples, BPCAfill will efficiently use all the provided informations and output the best result, even when there are large number of missing entries.
In addition, it is very easy to use, because it do not need any type of hand tuning parameters.
See also the paper,
Oba, S., Sato, M., Takemasa, I., Monden, M., Matsubara, K., and Ishii, S. A Bayesian Missing value estimation method, Bioinformatics, (2003), to appear.
13 May. 2003 : first release of the Java version
MATLAB version is hereJar file JBPCAfill.jar (170kb)
Put the JAR file in arbitrary directory.
Prepare a data file.

Open the JBPCAfill application frame from command line.
> java -classpath JBPCAfill.jar JBPCAfill
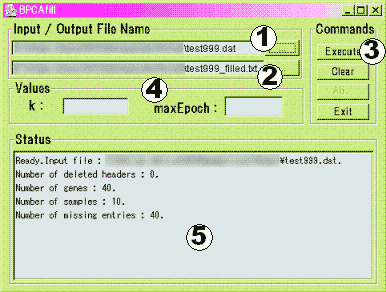 (1) Set the input filename.
(1) Set the input filename.
(2) Set the output filename. ( When the input filename (ex. test999.dat) is set, default filename (ex. test999_filled.dat) is set )
(3) Click the EXECUTE button, and the calculation starts.
Please wait for it to converge.
It will take some minutes or hours till finish.
When it finished, a new output file is written.
Missing entries will be filled by the estimated values.
(4) (OPTIONAL) If you want to handle, extraordinarily large data set, you may set some parameters, and you can obtain coarse estimation in short calculation time.
But, the default is the most recommended.
(5) Something appears in this window, but, please don't care.
> java -classpath JBPCAfill.jar JBPCAfill sample999.dat sampleFilled.dat