NeuroMorphoKit-K: A multiphysics simulation environment for neuromorphological dynamics
Overview
"NeuroMorphoKit" is a distributed simulation framework which captures three different physical layers including reaction-diffusion processes, membrane dynamics, and F-actin-based cytoskeletal kinetics. The software is developed under the Next-Generation Integrated Simulation of Living Matter (ISLiM), Computational Science Research Program, RIKEN.
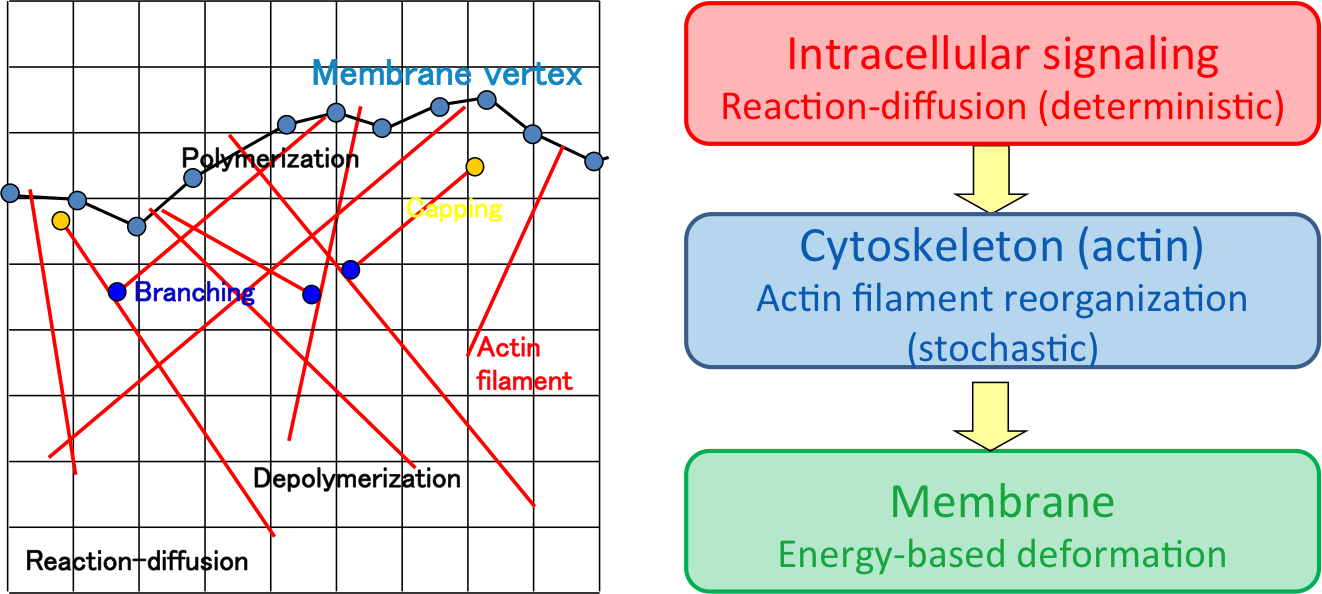
The current version of the NeuroMorphoKit provides C++ code, which is optimized in the K computer of the RIKEN Advanced Institute for Computational Science, for cellular morphology related simulation elements including efficient computation of stochastic kinetic events of actin filaments, deformation simulation of plasma membrane based energy optimization in the presence of extracellular obstacles, and simple reaction-diffusion solver for arbitrary reaction species in two-dimensional environments based on the paper:
Nonaka, S., Naoki, H. and Ishii, S. (2011)
A multiphysical model of cell migration integrating reaction–diffusion, membrane and cytoskeleton.
Neural Networks, 24, pp.979-989. (publisher's website)
Availability
Source code of NeuroMorphoKit is available upon request. Please contact using the e-mail address below.
License
Software requirements
- Library tested with Version
- GSL - GNU Scientific Library 1.15 (manually build on K-1.2.0-07)
- zlib 1.2.3 (on K-1.2.0-07)
Installation instructions can be found in the README.txt file in the root directory of the source code package.
Software contents
Main program
cell is a single binary executable file which is build from the source code package. The general behavior of cell including simulation settings and parameter values are configured by three input files listed below.
Configuration files for simulation
- File Description
- systeminfo.dat Simulation settings which include definition of simulation space, initial number of actin filaments, simulation time step/duration, and some physical constants.
- param.dat Physical and chemical constants of the simulation related biophysical phenomena from the published scientific literature.
- unknownparam.dat Physical and chemical constants of the simulation related biophysical phenomena.
Job script
run.sh is a sample job script for the K computer under the simulation assumption where short actin filaments are equally distributed near the plasma membrane surface and extra-cellular directions in the initial condition.
Extended simulation models
Two extended versions of the simulation model which incorporates 1) filament-filament binding by an actin cross-linker Fascin and the elastic properties of actin filaments based on a spring network model and 2) 3D plasma membrane deformation will be released separately in March 2013.
References
Nonaka, S., Naoki, H. and Ishii, S. (2011)
A multiphysical model of cell migration integrating reaction–diffusion, membrane and cytoskeleton.
Neural Networks, 24, pp.979-989.